Eukaryotic Cytosolic Ribosomes
Background
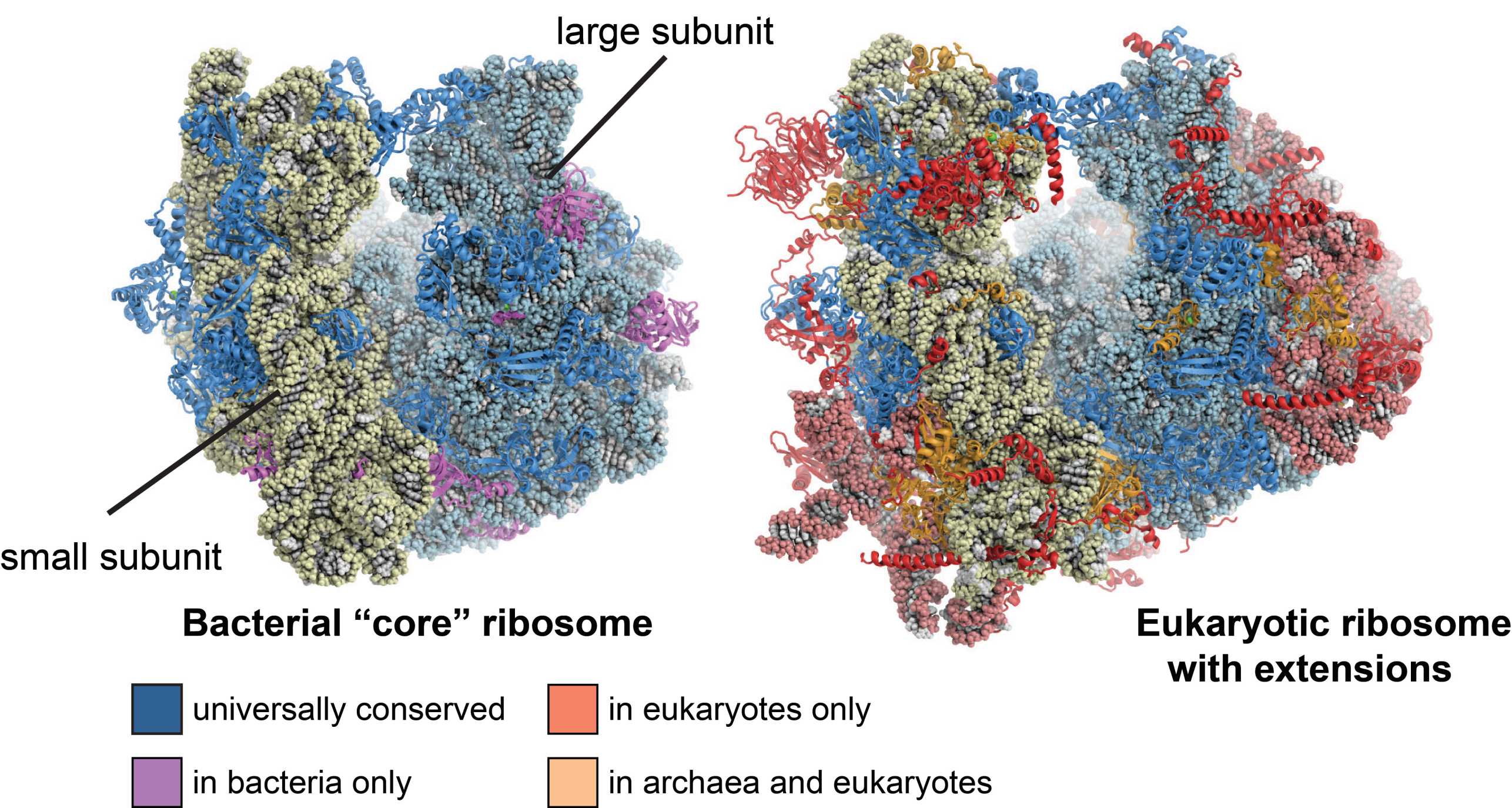
Ribosomes are large molecular machines the catalyze the translation of genetic information into proteins, one of the most fundamental processes in every organism. They consist of a small and a large subunit, both of which are composed of ribosomal RNA (rRNA) and ribosomal proteins. The atomic X-ray structures of bacterial ribosomes that emerged in the early 2000s helped understanding many fundamental aspects of the translation process and provided fascinating new functional insights into bacterial translation in particular. In 2009, this pioneering work was awarded with the external page Nobel Prize in Chemistry.
However, at that time, only little was known about the more complex structure and function of eukaryotic cytosolic ribosomes, which are significantly larger than bacterial ribosomes: While bacterial ribosomes have a molecular mass of 2.6MDa, eukaryotic ribosomes are almost twice as large with a molecular mass of 4.3MDa, contain extra rRNA and 45 additional ribosomal proteins, some of which are known to be involved in signaling. This increase in the number of molecular components is an adaptation to the more complex organization of the eukaryotic cell and is also reflected by the far more sophisticated machinery for translation initiation, protein targeting and ribosome biogenesis.
Consequently, several structural biology labs focused on solving the structure of the eukaryotic cytosolic ribosome by means of X-ray crystallography, an effort that resulted a decade later in the first atomic structures of the eukaryotic small and large subunits in complex with initiation factors from the ciliate Tetrahymena thermophila determined by our lab (external page Rabl et al. 2011, external page Klinge et al. 2011) and of the yeast ribosome solved by the Yusupov group (external page Ben-Shem et al. 2011).
Since then, we have been investigating a number of functional complexes of the eukaryotic ribosome, first by X-ray crystallography and more recently by cryo-EM, since the “resolution revolution” in the cryo-EM field allows visualization of structures at atomic detail. The structures include initiation (external page Weisser et al. 2013, external page Erzberger et al. 2014, external page Aylett et al. 2015, external page Quade et al. 2015) and targeting (external page Kobayashi et al. 2018) complexes, as well as structures of ribosome assembly intermediates (external page Greber et al. 2015, external page Scaiola et al. 2018).
Eukaryotic Cytosolic Translation Initiation
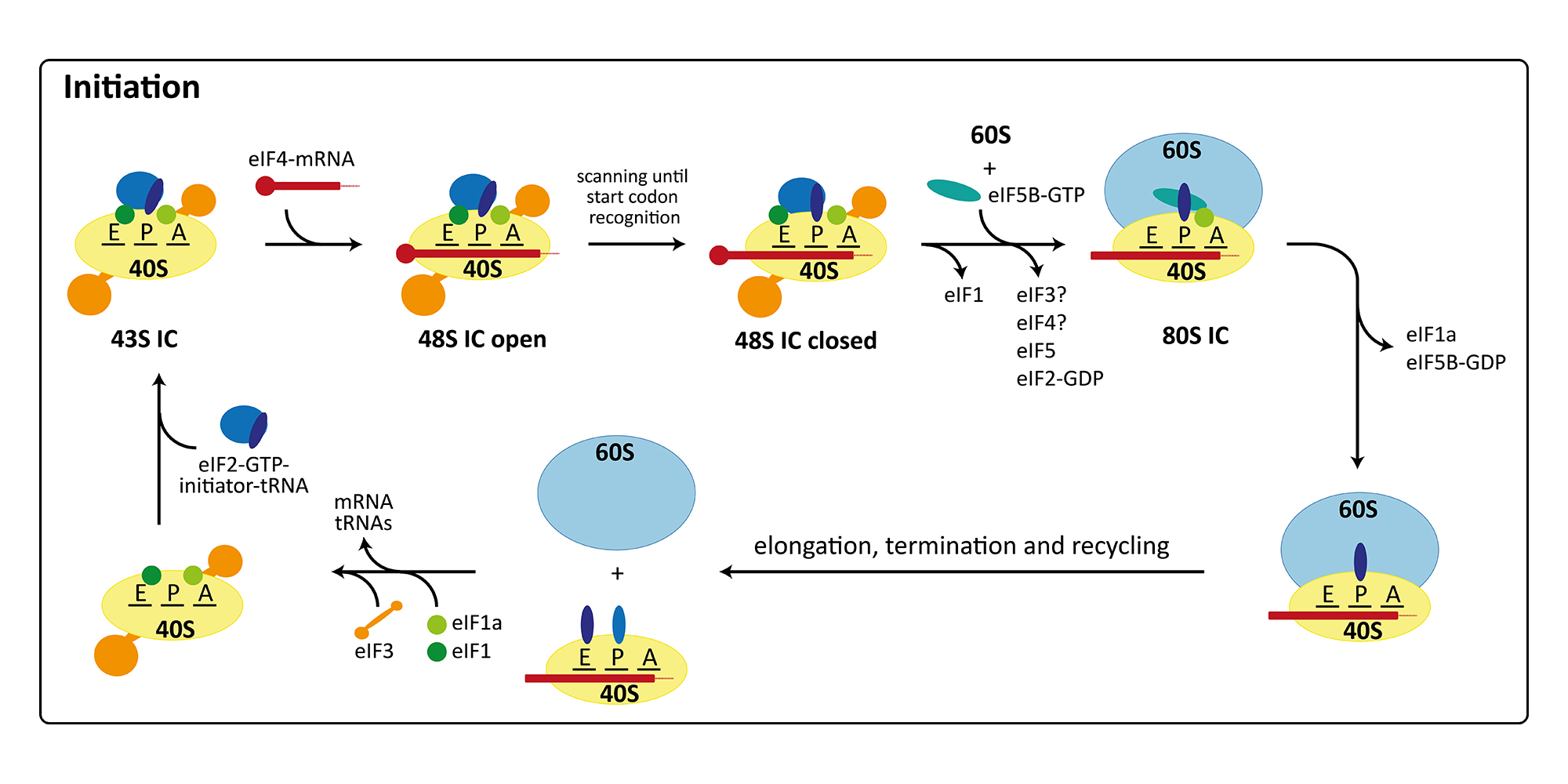
The initiation of protein synthesis in eukaryotes involves at least twelve factors, compared to only three factors required for initiation in bacteria. This is mainly due to the more elaborate mRNA structure possessing a 5’-cap, a 3’-poly(A)-tail as well as long 5’-untranslated regions with secondary structure elements. Therefore, initiation factors are involved in binding and unwinding of the mRNA, in scanning of the 48S pre-initiation complex (PIC) for the start codon, and finally in determining the correct open reading frame. Furthermore, translation initiation is the target of regulation in a number of cellular processes including development, differentiation, stress response, and neuronal function. Consequently, many diseases, including cancer and metabolic disorders, are connected with improper functioning or regulation of the initiation of protein synthesis. Additionally, the cellular translation machinery can be hijacked by some viruses for their own reproduction. In these cases, the ribosomes of the host organism directly bind to a secondary structure element in the viral mRNA termed internal ribosomal entry site (IRES), which allows initiation with a reduced set of initiation factors or entirely without the canonical initiation machinery. In our group, we are currently focusing on the canonical initiation process (external page Aylett et al. 2015, external page Erzberger et al. 2014) and on the non-canonical processes occurring during IRES mediated initiation (external page Quade et al. 2015) or re-initiation (external page Weisser et al. 2017).
Structural Insights into Human Re-initiation Complexes
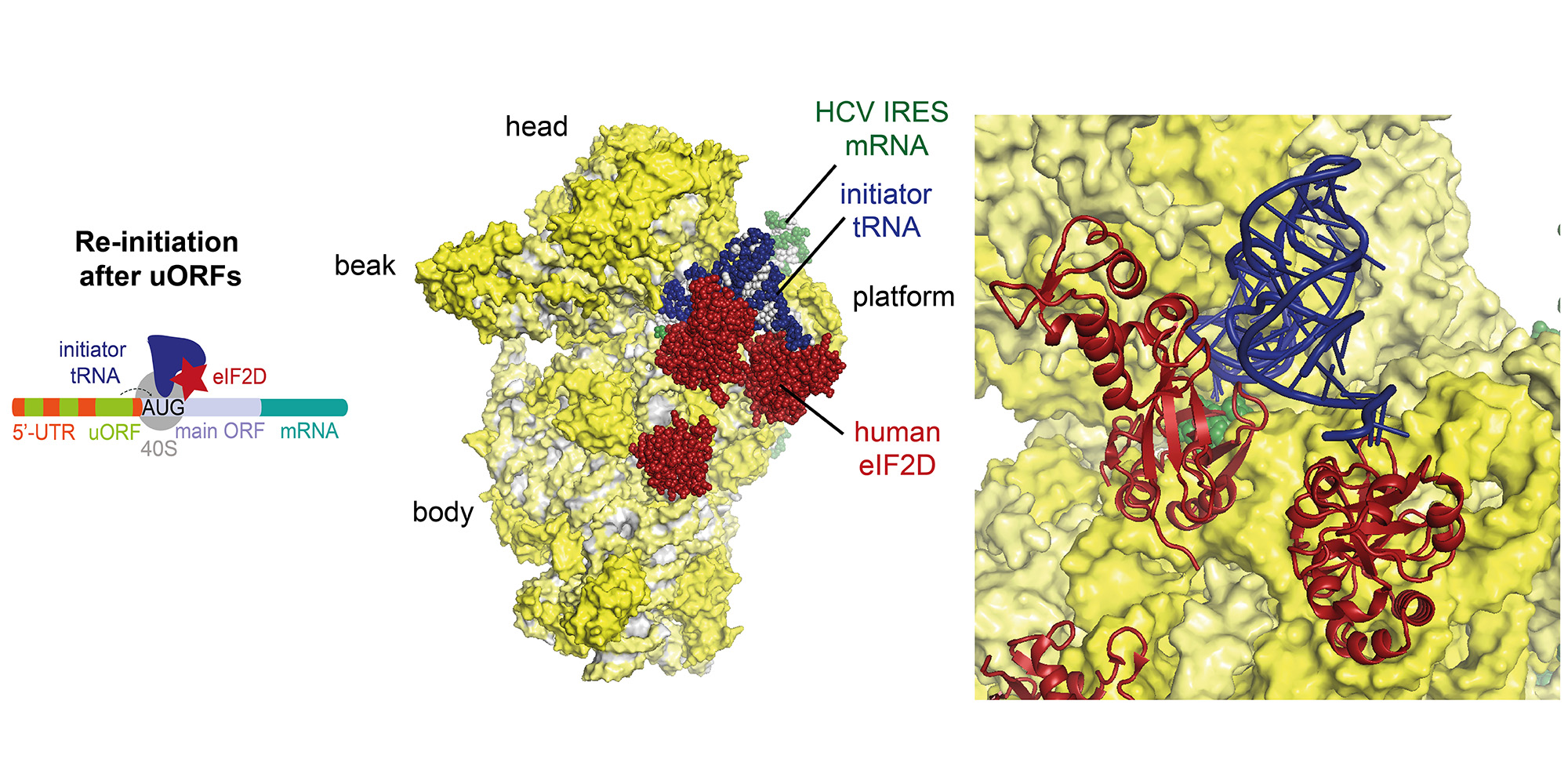
Re-initiation is a non-canonical process occurring during translation of mRNAs with short upstream open reading frames (uORFs), which regulate the translation through re-initiation of the downstream main ORF. Re-initiation is facilitated by non-canonical factors such as eIF2D or MCT-1/DENR.
Recently, we were able to determine the structures of re-initiation factors in complex with the small ribosomal subunit, mRNA and initiator tRNA by a combined approach using X-ray crystallography and single-particle cryo-EM (external page Weisser et al. 2017). In particular, we observed in our structure that ribosomes can re-initiate translation on the same mRNA by three distinct interactions of eIF2D (or the homologous complex of MCT-1/DENR) with the initiator tRNA and 40S subunit emulating the function of several canonical translation initiation factors by competing for the same binding sites.